Every cell in our body contains the same DNA sequence. So, if the order of the nucleotides is the same, how does cell specialization arise? The answer lies in how the DNA is folded in a cell, exposing certain regions for expression while repressing others. Following discoveries of chromosomes in 1879 by Fleming, Thomas Hunt Morgan’s establishment of chromosomes harboring the developmental program in Drosophila and identification of DNA as the primary carrier of genetic information (see this blog post), many researchers sought to investigate the concept of genetics. In 1930, H.J. Muller provided evidence of heritable changes in phenotype without any changes in genes [2]. This along with observations of the divergence of cell phenotypes and the subsequent maintenance over dividing cells indicate that there are inheritable changes in gene expression independent from alternation of the DNA sequence termed “epigenetics”.
In this blog series, I will provide an overview of different epigenetic changes that could occur, their mechanisms, technological techniques used to investigate them and, their association with disease.
Chromatin dynamics
Before going into different types of epigenetic changes that can occur, it is important to understand how chromatin is organized in a cell. Each human diploid cell contains approximately two meters of DNA making it necessary for a form of organization that would allow packaging within the confines of the tiny cell nucleus. The organization is mediated by small, positively charged proteins called histones that tightly wrap portions of the negatively charged DNA into structures called nucleosomes allowing for the DNA to condense in such a way such that they form chromatin (Figure 1). There are five different types of histones (H1, H2A, H2B, H3, H4) and eight of them (two each of H2A, H2B, H3, H4) form a nucleosome. The H1 histone acts as a “linker”, binding the entry/exit sites of DNA and sitting on top of the core eight histone structure of a nucleosome and acts as a stabilizer [3].
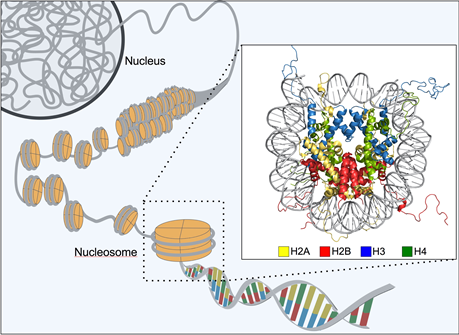
Figure 1: Organization of chromatin. Adapted from the OIST.
Chromatin and epigenetics
But, why are nucleosomes important? The distribution of nucleosomes in chromatin play a significant role in gene regulation by affecting which regions of DNA are accessible. Chromatin accessibility is the degree in which nuclear macromolecules like transcription factors are able to physically associate directly with DNA [4]. Regions with closed chromatin have tightly spaced nucleosomes which prevent transcriptional and regulatory machinery such as transcription factors from accessing the DNA directly. Open chromatin allows for genes within the inter-nucleosome regions to be expressed since the recognition regions are exposed for binding of the transcription factors (Figure 2). As we saw before, the expression of these genes can dictate cell types.
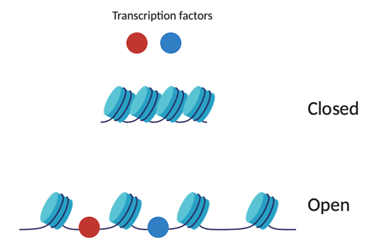
Figure 2: Schematic of open and closed chromatin. Created on BioRender.
Epigenetic changes occur on histones, DNA and through expression of something called non-coding RNAs (ncRNAs). Figure 3 shows examples of different types of epigenetic modifications that can occur, and we will go more into detail about them and different mechanisms in this blog series.
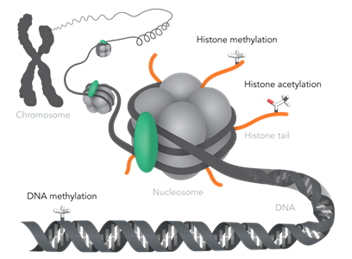
Figure 3: Examples of epigenetic modifications. Adapted from Horizon Discovery.
Summary
Chromatin is an overarching term that takes into account DNA in association with proteins where nucleosomes are the basic structural and functional unit. Epigenetic modifications like methylations and acetylation can occur on DNA and/or histones. These epigenetic modifications can regulate gene expression without changing the genetic sequence directly by altering which regions of chromatin are open and closed.